Back to top
miCORE Amplicon Metagenomics

Uncover microbial diversity with targeted amplicon sequencing.
Amplicon metagenomics enables precise analysis of bacterial and fungal communities by focusing on taxonomic marker regions such as 16S rRNA and ITS. Whether comparing experimental groups, monitoring shifts, or characterizing unknown communities, this method provides accurate and efficient insights into microbial diversity.
What You Can Achieve
- Explore bacterial, fungal, or archaeal community composition
- Compare taxonomic shifts across experimental conditions
- Detect nutrition-dependent microbiome changes (e.g., gut microbiota)
- Characterize rare or environment-specific taxa (e.g., deep-sea archaea, airborne fungi)
- Monitor community dynamics in bioreactors and other controlled systems
- Optimize future sampling strategies with high-resolution data
Before You Start
To set up your amplicon metagenomics project effectively, consider:
- Scientific objective – survey, experimental comparison, or functional inference?
- Starting materials – DNA, environmental sample, sewage sludge
- Desired services – from DNA extraction to full bioinformatics analysis
- Result type – raw data only, or full bioinformatics reporting
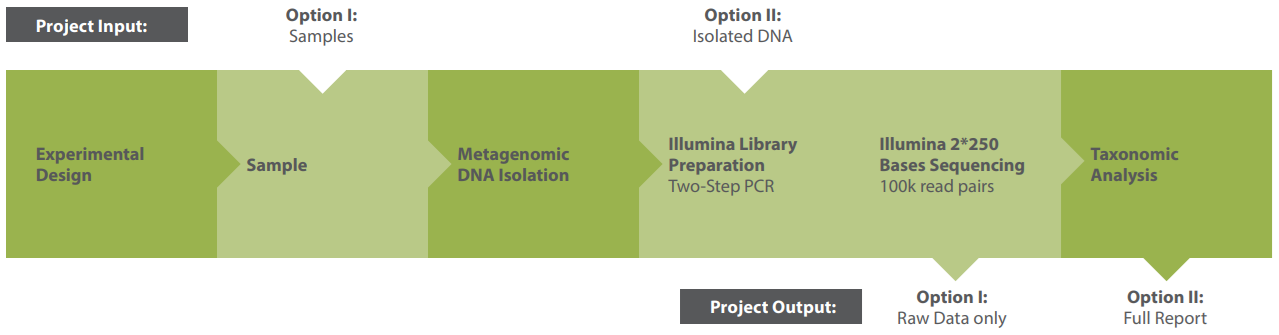
Modular Workflow
Outsource the complete workflow or select specific modules. Our process is designed for flexibility. Typical steps include:
Raw data includes:
- Sequencing quantity and quality assessment (.xlsx)
- Raw sequencing data (.fastq per sample)
- Project summary report (.pdf)
Bioinformatics Analysis
Standard Bioinformatics
Our standard 16S/ITS analysis module provides:
- Comprehensive interactive report (.html)
- Amplicon Sequence Variants (ASVs / zOTUs) in .fasta format
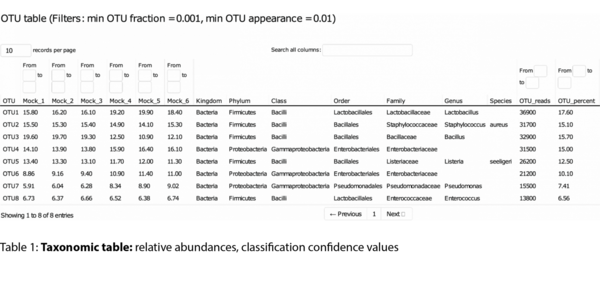
- ASV abundance, taxonomy, prediction confidence (.tsv, .xlsx)
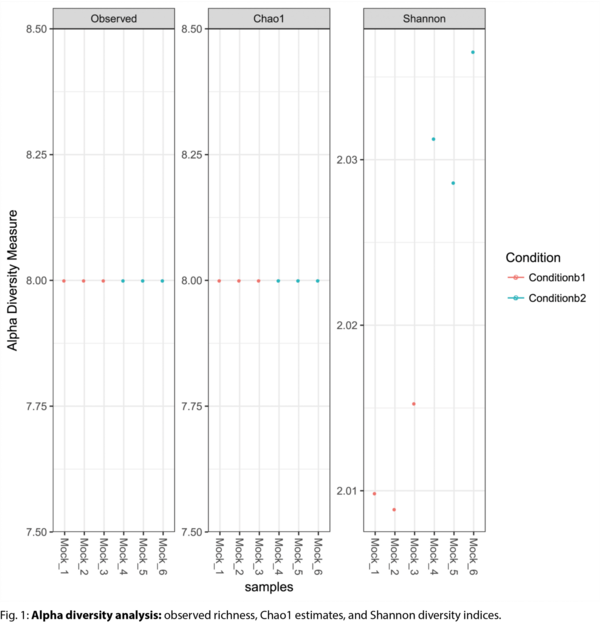
- Intrasample diversity & rarefaction curves (alpha diversity) (.tsv, .pdf)
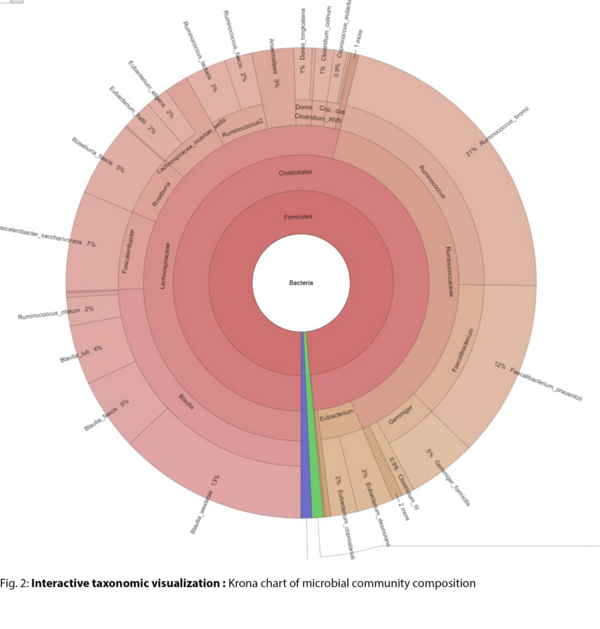
- Interactive Krona charts for taxonomic exploration (.html)
Complementary Bioinformatics (optional, additional costs)
- Comparative Analysis
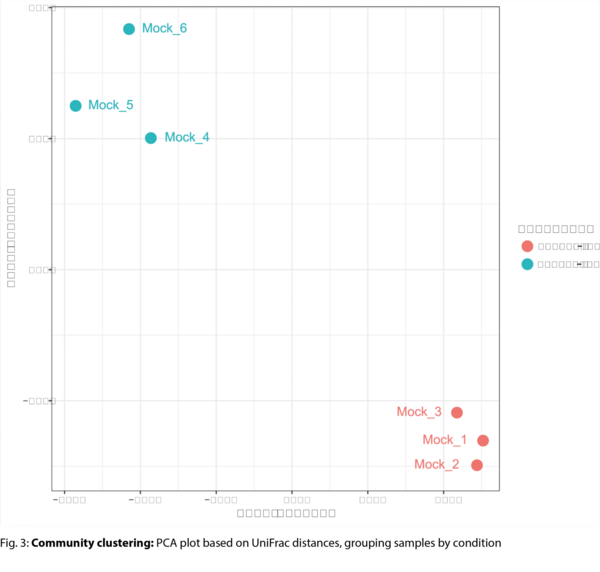
- Inter-sample diversity & clustering (beta diversity) (.tsv, .pdf)
- Principal Component Analysis (PCA) of sample relationships (.tsv, .pdf)
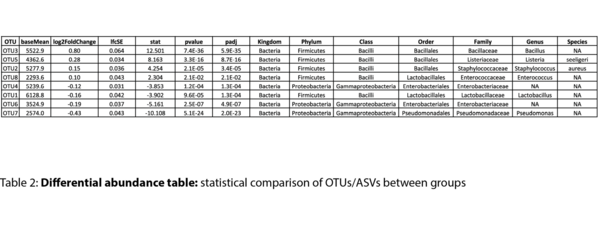
- Differential ASV analysis with statistics (.tsv, .html)
- Sample correlations (.tsv, .pdf)
- Visual summaries of statistical results
- Functional Profiling
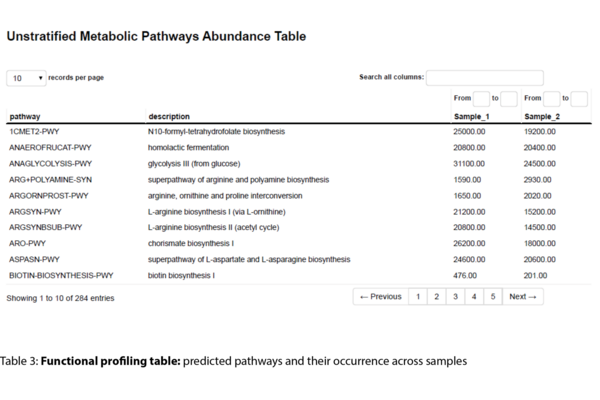
- Prediction of functional profiles based on identified taxa (.tsv, .html)
Example Results
All results are delivered in publication-ready figures and interactive formats, providing both high-level overviews and nucleotide-level detail.
Turnaround Time
- Sequencing only: 15 working days
- DNA isolation: +5 working days
- Bioinformatics analysis: +3 working days
- Express service available upon request
Sample Requirements
- See User Guide